Basic Operation
Quick LinksThis tutorial provides a broad overview of how to use ArVirInd (Arbo-Viral proteins from India and its neighbourhood) database to analyze and display data. ArVirInd database includes antigenic protein sequences from
Indian subcontinent countries included in ArVirInd database are:
India
Bangladesh
Bhutan
Maldives
Nepal
Pakistan
Sri Lanka
What are arboviral diseases?
“Arbovirus,” short for arthropod-borne virus, doesn’t refer to one particular virus. Rather, it refers to a type of virus transmitted via insects that bite and feed on blood. Arboviral disease is a general term used to describe infections caused by a group of viruses spread to people by the bite of infected arthropods (insects) such as mosquitoes and ticks. These infections usually occur during warm weather months, when mosquitoes and ticks are active.Arbo-viral Diseases included in ArVirInd database are:
Dengue viruses (all serotypes)
Japanese encephalitis
West Nile
Chikungunya
Chandipura etc.
What is antigenic proteins?
“Antigens” are molecular structures on the surface of viruses that are recognized by the immune system and are capable of triggering one kind of immune response known as antibody production. The term antigen originally referred to a substance that is an antibody generator. Antigens can be proteins, peptides (amino acid chains), polysaccharides (chains of monosaccharides/simple sugars), lipids, or nucleic acids.As stated above, antigens are molecules which are present on the surface of a pathogen. Their primary role is to elicit the response of the immune system. This in turn stimulates the production of antibodies, which are Y-shaped proteins that help fight off the infection.
A molecule that is capable of binding to an antibody or to an antigen receptor on a T-cell, especially one that induces an immune response.
What is b-cell epitope?
B-cell epitopes can be defined as a surface accessible clusters of amino acids, which are recognized by secreted antibodies or B-cell receptors and are able to elicit cellular or humoral immune response. A B-cell epitope is the antigen portion binding to the immunoglobulin or antibody.Adaptive immunity is mediated by T-cells and B-cells, which are immune cells capable of developing pathogen-specific memory that confers immunological protection. Memory and effector functions of B-cells and T-cells are predicated on the recognition through specialized receptors of specific targets (antigens) in pathogens. More specifically, B-cells and T-cells recognize portions within their cognate antigens known as epitopes. There is great interest in identifying epitopes in antigens for a number of practical reasons, including understanding disease etiology, immune monitoring, developing diagnosis assays, and designing epitope-based vaccines.
Identification of B-cell epitopes is a fundamental step for development of epitope-based vaccines, therapeutic antibodies, and diagnostic tools. Epitope-based antibodies are currently the most promising class of biopharmaceuticals. ArVirInd will be useful in the study of immuno-informatics, diagnostics and vaccinology for arboviruses.
ArVirInd is available publicly via Internet at http://www.arvirind.co and in starts up, the Main Application Window appears with several different panes like Menu bar, Search Bar, Peptide Search option and important links.

To start let us search for some data in the ArVirInd databases.
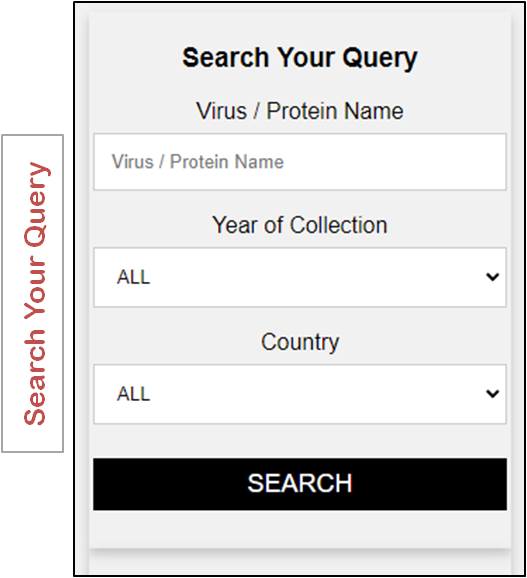 |
ArVirInd homepage provides the user the following ways to search:
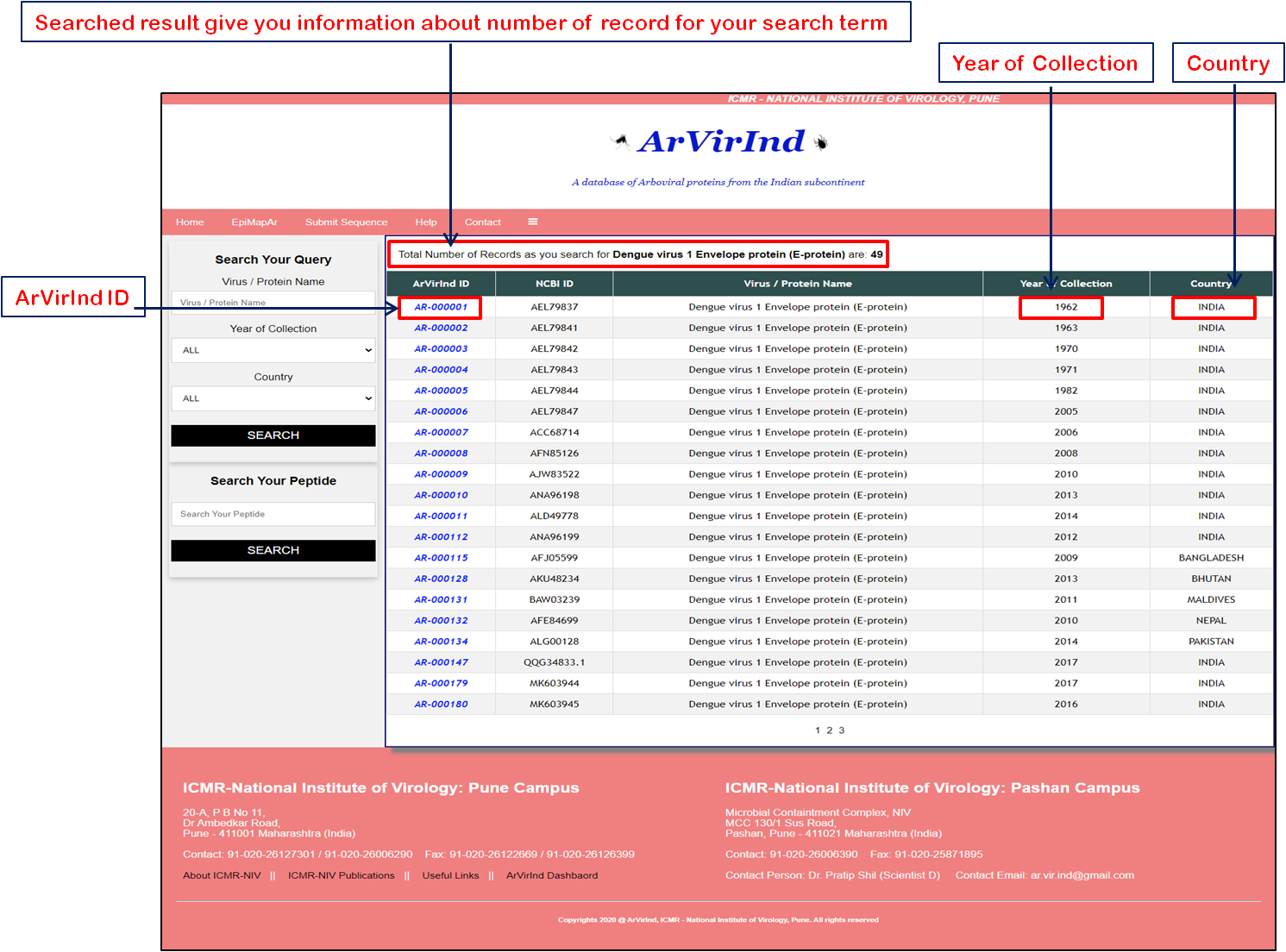
a) Search by “Virus Name”, b) search by “Country of Origin”, c) search by "Year of Origin" or a combination of these options. This is a unique feature of the database. This search option is facilitetes with auto-search and auto-suggest query for user. Let us now search for the virus "Dengue virus 3" in the ArVirInd database. To do this, follow the steps below:
A query returns a tabular output which gives the list of all entries in the searched category. From this table user can choose any particular entry by clicking on the respective "ArVirInd ID". |
ArVirInd also allows the user to search the database by submitting a query peptide (or epitope) through the "Search Your Peptide" search box
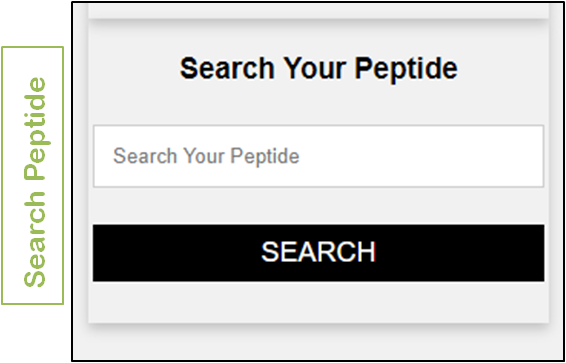 |
Let us now "search for peptide" as "NEMVLLT" in the ArVirInd database. To do this, follow the steps below:
A query returns a tabular output which gives the list of all entries highlighted with searched peptide term. From this table user can choose any particular entry by clicking on the respective "ArVirInd ID". From results user also came to know about how many sequence records in the database have searched peptide. |
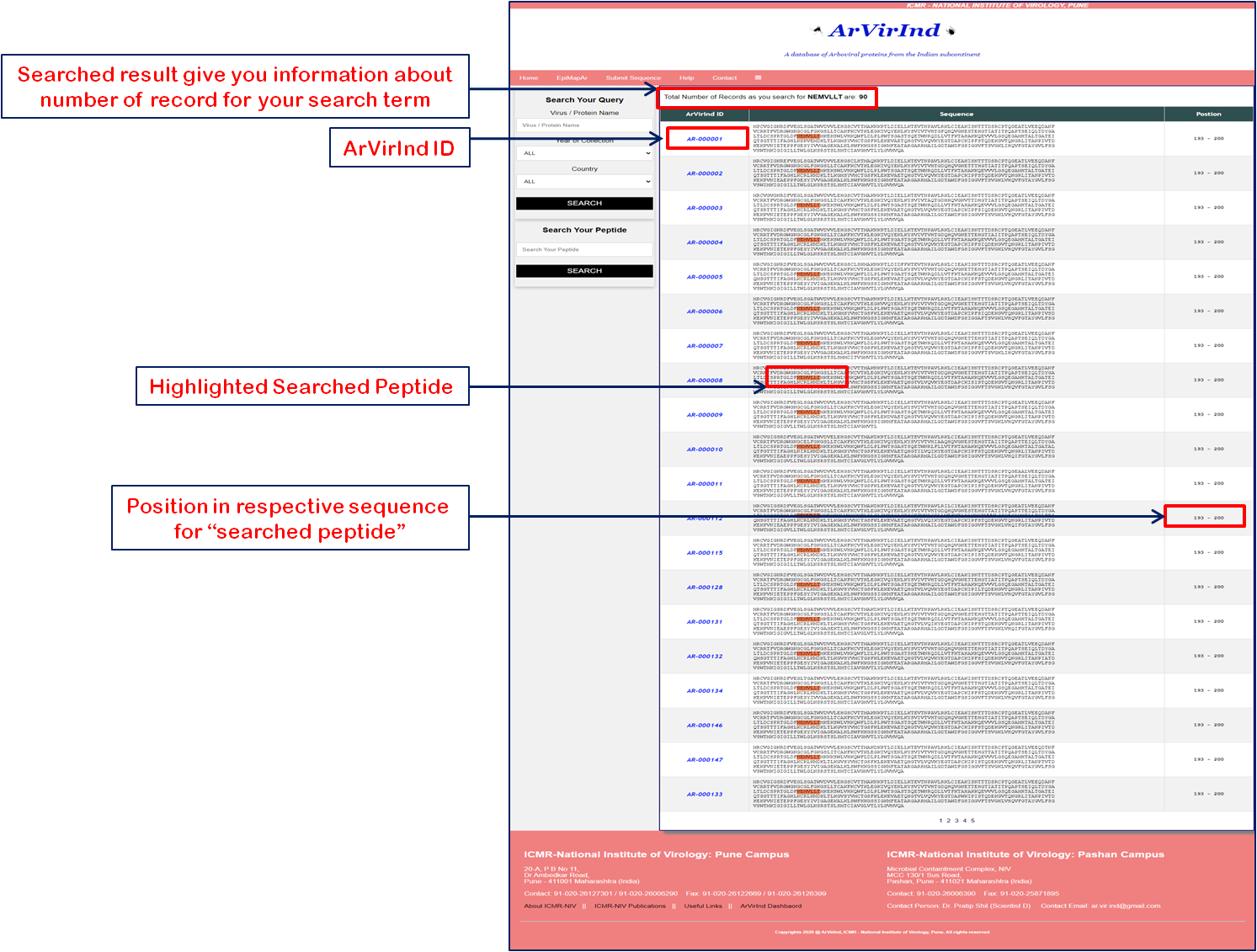
Once the user select the particular ArVirInd ID and respective record is loaded, the main window should look somewhat like the image below. Each ArVirInd record gives you the details as below:
- ArVirInd record Identifier
- Respective ArVirInd ID
- Respective NCBI id
- Virus Protein Name
- Protein
- Virus Family
- Host
- Year of Collection
- Amino acid sequence
- Vaxigen Score
- B-cell epitope
- T-cell epitope
- N-glycosylation site
- Related PDB id
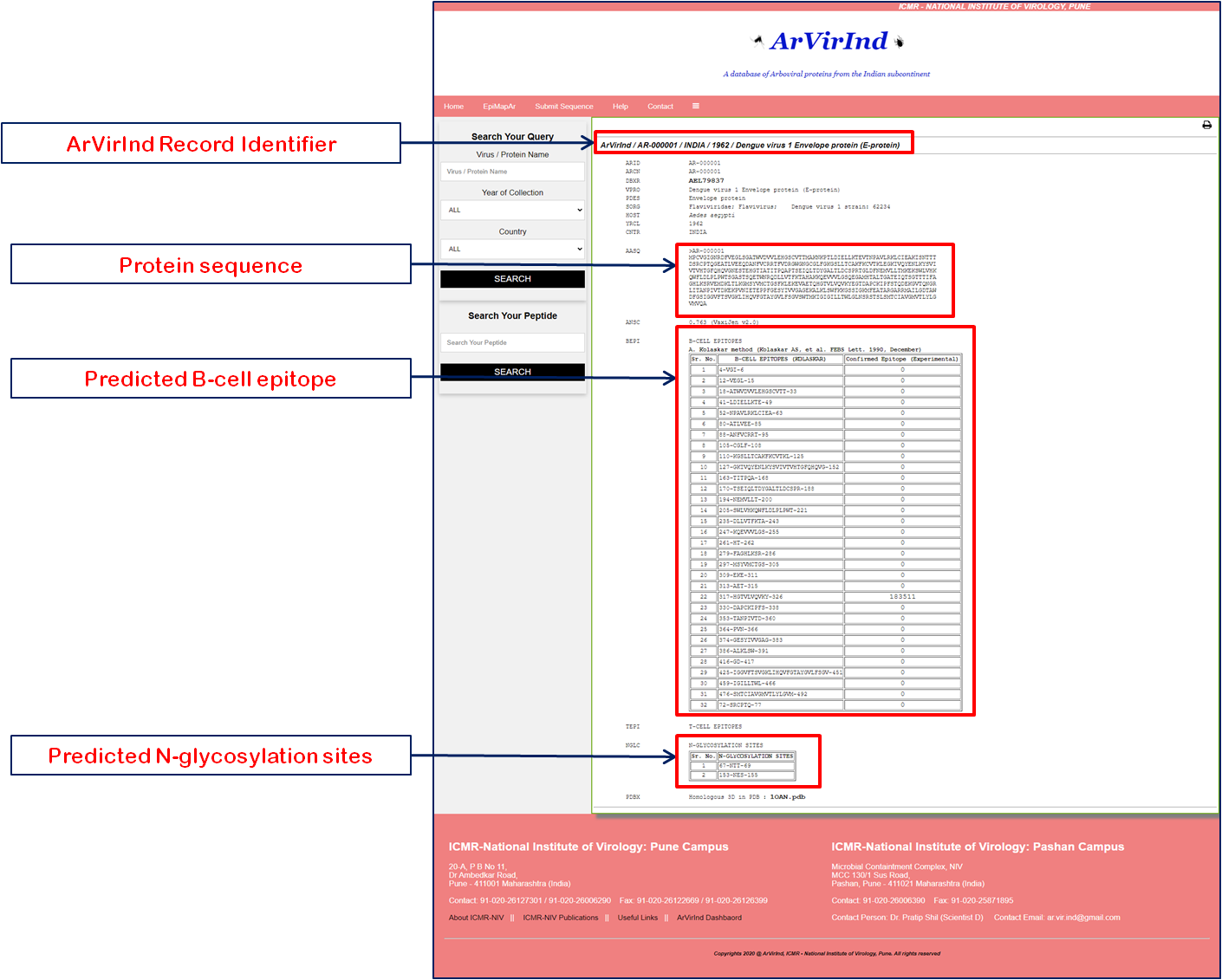
For detail about the ArVirInd file format please >>>> CHECK HERE
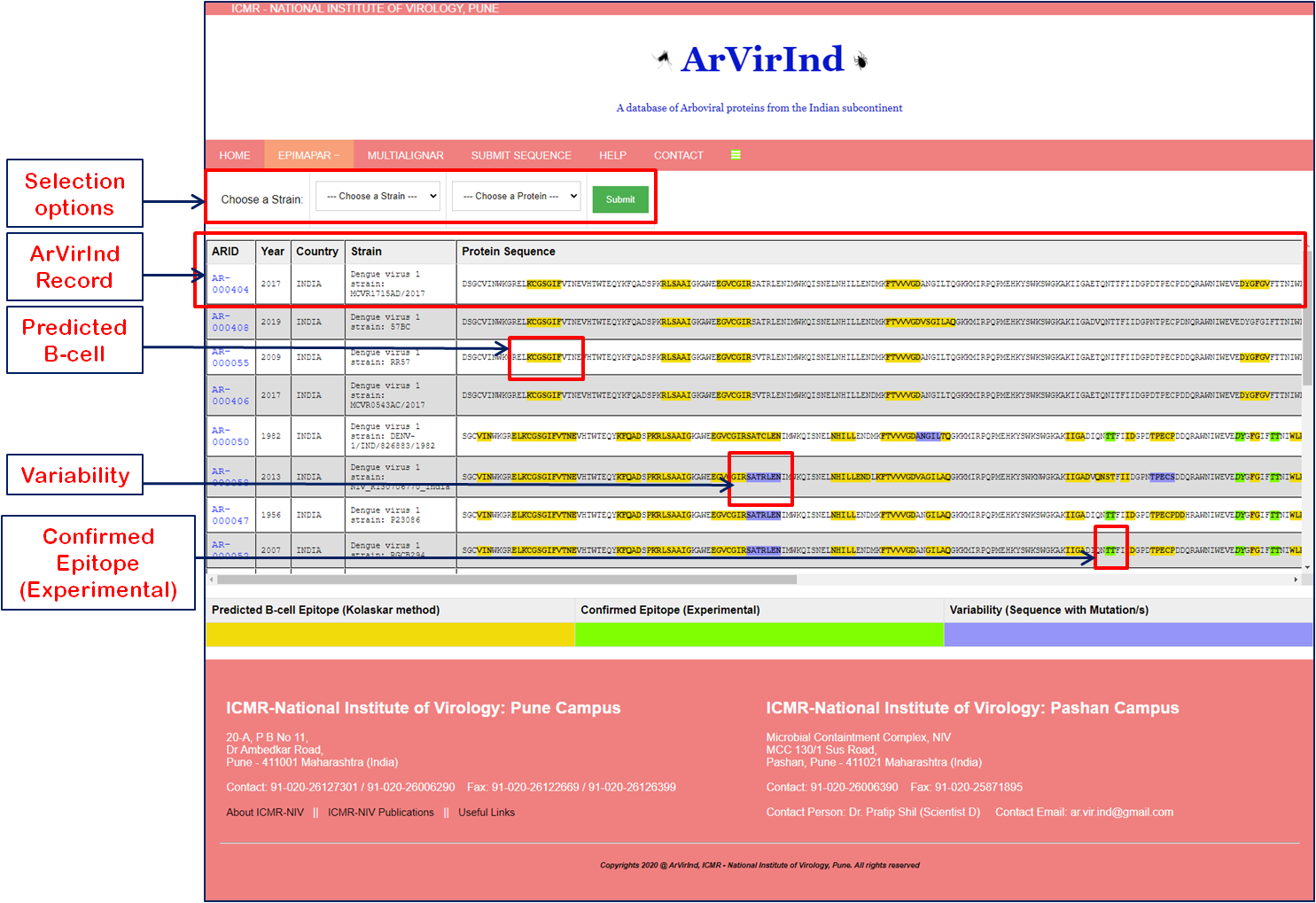
The ArVirInd database has an epitope mapping tool called “EpiMapAr”, which helps display the predicted and known B-cell epitopes (antigenic regions) on the database sequences.
This tool can be accessed on the “EpiMapAr” page. This gives the user the choice of “virus” and “antigenic protein”.
As an output, it displays all the available sequences in the database for the selected category as below
- Yellow highlighted sequence pattern represent Predicted B-cell Epitope (Kolaskar method)
- Green highlighted sequence pattern represent Confirmed Epitope (Experimental).
- Purple highlighted sequence pattern represent Variability (Sequence with Mutation/s).
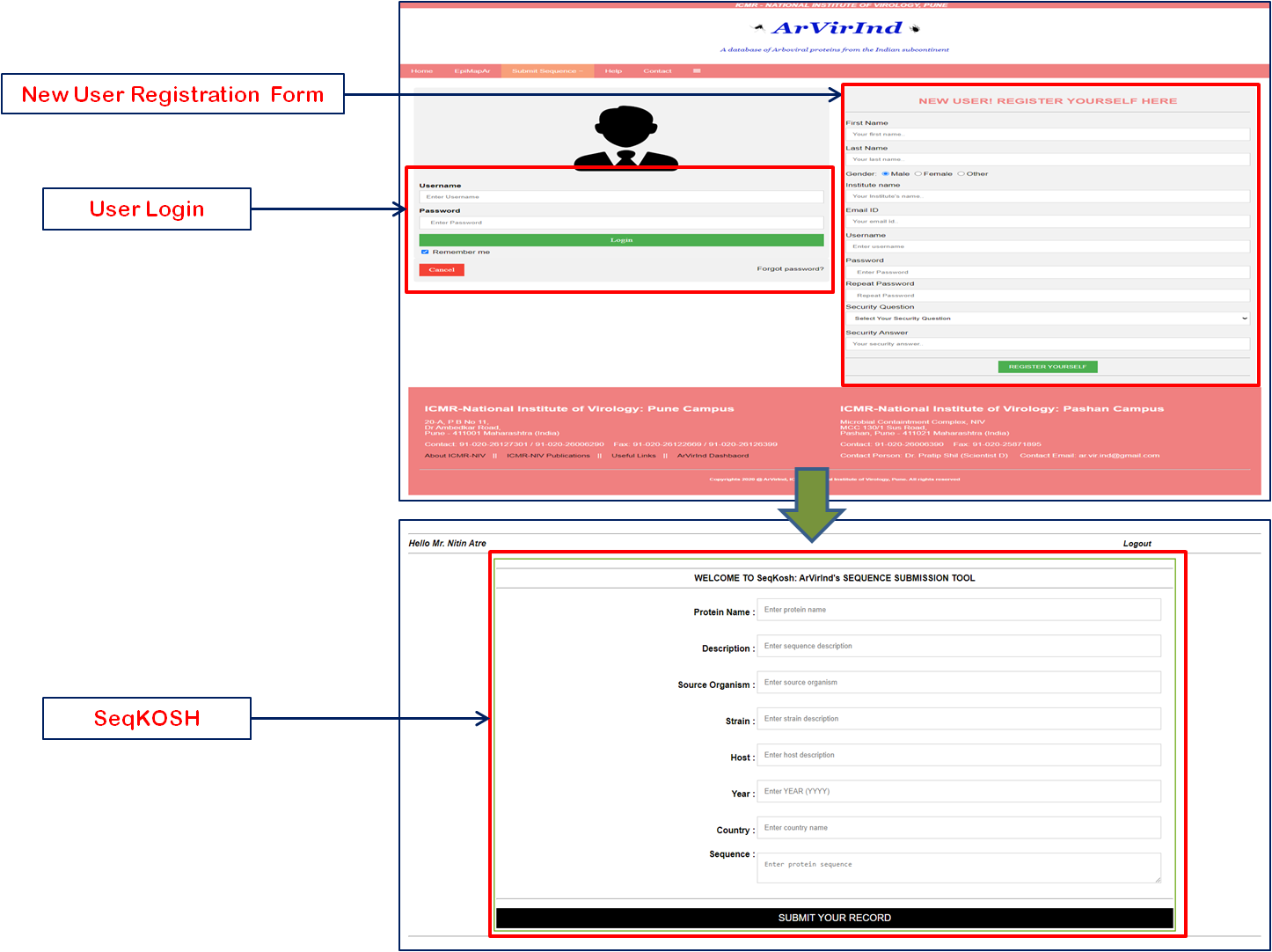
The ArVirInd database, provides an opportunity for the user to submit sequences of viral strains isolated from emerging outbreaks of Dengue, Chikungunya, West Nile, Japanese encephalitis, and Chandipura.
The sequence submission tool is enlisted on the submission page.
The new user has to register with a valid institutional email id. This will be verified by our team before confirmation of registration.
After successfully registration, the user can log in and submit the sequences using our sequence submission tool called, SeqKosh.
All user-submitted sequences will be processed, analyzed, annotated, and uploaded by the ArVirInd team.
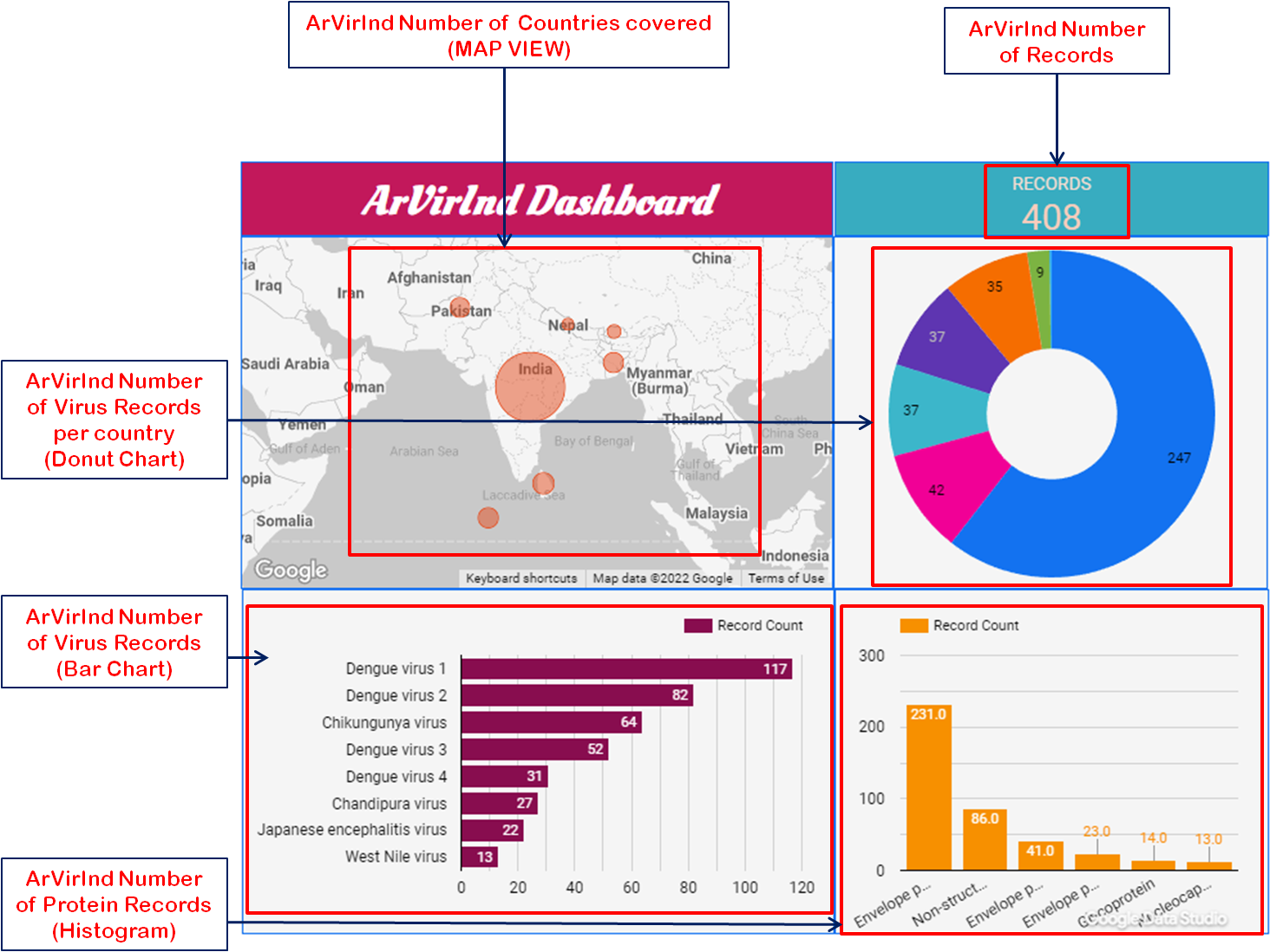
The ArVirInd database also provides, an interactive dashboard available at http://arvirind.co.in/dashboard/.
These summarises, the country-wise, virus-wise, and protein-wise records count information about the ArVirInd database.
A database, named ArVirInd, is created as a repository of information on curated antigenic proteins. This enlists sequences by country and year of outbreak or origin of the viral strain. For each entry, antigenic information is provided along with functional sites, etc.
Researchers can search this database by virus/protein name, country, and year of collection (or in combination) as well as peptide search for epitopes. It is available publicly via the Internet at http://www.arvirind.co.in. We anticipate that this database will be maintained and constantly expanded with information added for new and emerging strains of arboviruses.
ArVirInd will be useful in the study of immune informatics, diagnostics, and vaccinology for arboviruses. We believe that the ArVirInd knowledge base will benefit researchers working in immuno-informatics, arboviral diagnostics, and vaccinology. ArVirInd has the potential to become an important source of information on arboviral proteins for researchers working in India as well as countries like Bhutan, Nepal, Maldives, Bangladesh, Sri Lanka, and Pakistan. The database will be updated and expanded periodically.